VCF to PED Converter
The VCF to PED converter allows you to parse a vcf file (specification) to create a linkage pedigree file (ped) and a marker information file, which together may be loaded into ld visualization tools like Haploview. There is both an online version of this tool and a perl script
Online version
You can access the online version of the converter tool from the tools link in the menu bar at the top of every page in the Ensembl GRCh37 browser.
The input interface of the online version looks like:
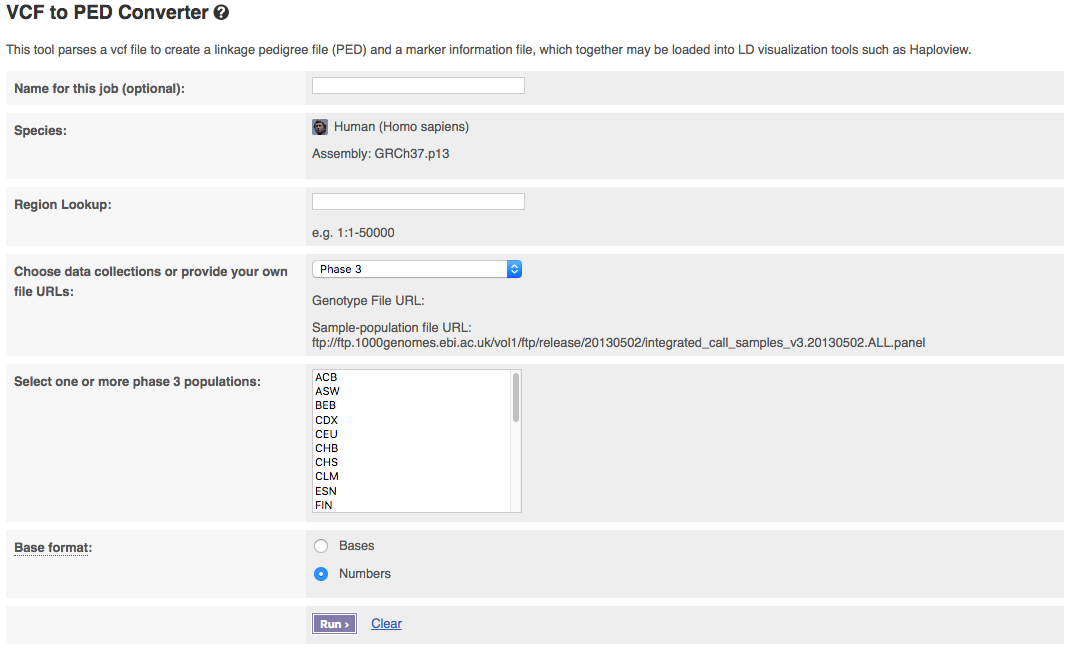
The tool allows you to pick which phase of the 1000 Genomes Project you want to get data from. If you have a publicly visible VCF file and corresponding sample-population mapping file, you could get data from these by selecting “Provide file URLs”.
Populations can be selected from the drop down list. To select multiple populations please hold the ctrl key (on windows/linux) or the cmd key (macs).
After clicking “Run” the system produces your final files:
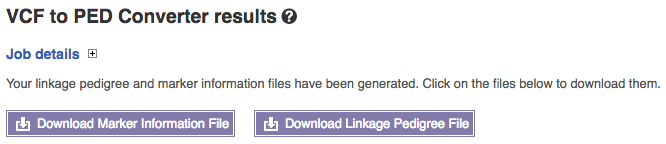
The marker information file and linkage pedigree file can be downloaded by clicking the links. In the linkage pedigree file the columns for father’s ID, mother’s ID, sex and affection status are all set to zero, signifying ‘unknown’.
API script
A perl API script version of the converter tool is available from the ftp site. You can also find a link to the script from either the tools link in the menu bar at the top of every page or from the manage your data link which is on the left hand menu of many pages in the browser.
This script converts locally or remotely accessible vcf files to linkage pedigree files. If the input file is only remotely accessible then it must be compressed by bgzip and indexed by tabix. There is no requirement to compress vcf files if they are held locally, but large files will be read more quickly using tabix. If the vcf file is compressed then you must have tabix installed.
The script is run from the command line and it takes the following arguments:
-vcf (required argument) Path to a locally or remotely accessible tabix indexed vcf file
-sample_panel_file (required argument) Path to a locally or remotely accessible sample panel file, listing all individuals (first column)and their population (second column)
-population (required argument) A population name, which must appear in the second column of the sample panel file. Can be specified more than once for multiple populations.
-tabix (optional argument) Path to the tabix executable. If the vcf file is compressed and this argument is not specified, the default is to search PATH for ‘tabix’
-output_ped (optional argument) Name of the output ped file. The default name is region.ped (e.g. 1_100000-100500.ped).
-output_info (optional argument) Name of the output info file (marker information file). The default name is region.info (e.g. 1_100000-100500.info).
-output_dir (optional argument) Name of a directory in which to put the output files.
| -base_format (optional argument) number | letter (defaults to number) if letter is specified the genotypes will be expressed as ATGC rather than 0123, by default this script uses the old style of plink allele annotation which used A => 1, C => 2, G => 3 and T => 4 |
-help (optional argument) Print out the help documentation
Here is an example of a command line for running the script:
perl vcf_to_ped_converter.pl -vcf ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20110521/ALL.chr13.phase1_integrated_calls.20101123.snps_indels_svs.genotypes.vcf.gz -sample_panel_file ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20110521/phase1_integrated_calls.20101123.ALL.sample_panel -region 13:32889611-32973805 -population GBR -population FIN